Code
from pyprojroot import here
import sys
python_dir = str(here("python"))
if python_dir not in sys.path:
sys.path.insert(0, python_dir)
%load_ext autoreload
%autoreload 2Erik De Luca
July 15, 2025
Import functions and data into the notebook. The data is located in the data folder, while the functions are in the python folder. To enhance the readability of the notebook and reduce code redundancy, certain functions are organized into python files and imported as needed.
# data manipulation
import pandas as pd
import numpy as np
import polars as pl
import torch
# data visualization
import matplotlib.pyplot as plt
import seaborn as sns
# models
from hmmlearn import hmm
from pyprojroot import here
import pyro
import pyro.distributions as dist
import pyro.distributions.constraints as constraints
from pyro.infer import SVI, TraceEnum_ELBO, config_enumerate
from pyro.optim import Adam
from scipy.stats import poisson
data = pd.read_csv(here("data/recent_donations.csv"))
data
# remove columns y_2020 to y_2023
# data = data.drop(columns=["y_2020", "y_2021", "y_2022", "y_2023"])| unique_number | class_year | birth_year | first_donation_year | gender | y_2009 | y_2010 | y_2011 | y_2012 | y_2013 | y_2014 | y_2015 | y_2016 | y_2018 | y_2019 | y_2017 | y_2020 | y_2021 | y_2022 | y_2023 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 26308560 | (1960,1970] | 1965 | 1985 | M | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 3 | 1 |
| 1 | 26309283 | (1960,1970] | 1966 | 2002 | M | 2 | 1 | 2 | 2 | 1 | 1 | 3 | 3 | 4 | 1 | 3 | 3 | 3 | 3 | 4 |
| 2 | 26317365 | (1960,1970] | 1961 | 1984 | M | 4 | 2 | 3 | 3 | 3 | 4 | 3 | 3 | 2 | 3 | 3 | 2 | 0 | 1 | 0 |
| 3 | 26318451 | (1960,1970] | 1967 | 1989 | M | 0 | 3 | 3 | 4 | 4 | 4 | 2 | 3 | 3 | 1 | 2 | 3 | 1 | 0 | 0 |
| 4 | 26319465 | (1960,1970] | 1964 | 1994 | F | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 9231 | 27220599 | (1970,1980] | 1980 | 2022 | M | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| 9232 | 27220806 | (2000,2010] | 2002 | 2022 | M | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 |
| 9233 | 27221247 | (1990,2000] | 2000 | 2022 | F | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| 9234 | 27221274 | (1960,1970] | 1966 | 2022 | F | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 |
| 9235 | 27221775 | (2000,2010] | 2004 | 2022 | M | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 |
9236 rows × 20 columns
# from pandas to polars
df = pl.from_pandas(data) # or pl.read_csv("file.csv")
# collect donation numbers along years
year_cols = sorted([c for c in df.columns if c.startswith("y_")])
T = len(year_cols)
obs = (df.select(year_cols)
.fill_null(0)
.to_numpy()
.astype(int)) # (N,T)
# prepare fixed covariates for pi
df = df.with_columns([
(pl.col("gender") == "F").cast(pl.Int8).alias("gender_code"),
((pl.col("birth_year") - pl.col("birth_year").mean()) /
pl.col("birth_year").std()).alias("birth_year_norm")
])
birth_year_norm = df["birth_year_norm"].to_numpy() # (N,)
gender_code = df["gender_code"].to_numpy() # (N,)
cov_init = np.stack([birth_year_norm, gender_code], axis=1)
# dynamic covariates for transition matrix
# normalize age
years_num = np.array([int(c[2:]) for c in year_cols])
ages = years_num[None, :] - df["birth_year"].to_numpy()[:, None]
ages_norm = (ages - ages.mean()) / ages.std()
# create dummy variable for covid years
covid_mask = np.isin(years_num, [2020, 2021, 2022]).astype(float)
covid_years = np.tile(covid_mask, (df.height, 1))
# A-covariate tensor (N,T,2)
cov_tran = np.stack([ages_norm, covid_years], axis=2)
# store in torch objects
obs_torch = torch.tensor(obs, dtype=torch.long)
cov_init_torch = torch.tensor(cov_init, dtype=torch.float) # (N,2)
cov_tran_torch = torch.tensor(cov_tran, dtype=torch.float) # (N,T,2)
print("obs :", obs_torch.shape) # (N,T)
print("pi covs :", cov_init_torch.shape) # (N,2)
print("A covs :", cov_tran_torch.shape) # (N,T,2)obs : torch.Size([9236, 15])
pi covs : torch.Size([9236, 2])
A covs : torch.Size([9236, 15, 2])The model uses two covariate blocks.
\(x^{\pi}_n = (\text{birth\_year\_norm},\;\text{gender\_code})\in\mathbb R^{2}\) affects the initial state,
while \(x^{A}_{n,t} = (\text{ages\_norm},\;\text{covid\_years})\in\mathbb R^{2}\) drives the transition at time \(t\).
The covariates on \(\pi\) are static covariates, while the covariates on \(A\) transition matrix are designed for dynamic changes to ensure model flexibility over the time.
Hidden state: \(z_{n,t}\in\{0,1,2\}\), reppresents the propensity to donate. Observed count: blood donations by donor \(n\) at the time \(t\), denoted as \(y_{n,t}\).
Priors for the intercepts
• \(\pi_{\text{base}}\sim\text{Dirichlet}(\boldsymbol{\alpha}_{\pi})\)
• \(A_{\text{base}}[k,\cdot]\sim\text{Dirichlet}(\boldsymbol{\alpha}_{A_k})\)
• \(\lambda_k\sim\text{Gamma}(2,1)\) for \(k=0,1,2\)
Slope parameters to be learned
• \(W_\pi\in\mathbb R^{K\times2}\)
• \(W_A\in\mathbb R^{K\times K\times2}\)
Initial-state distribution
\(\Pr(z_{n,0}=k\mid x^{\pi}_n)= \operatorname{softmax}_k\!\bigl( \log\pi_{\text{base},k}+W_{\pi,k}\cdot x^{\pi}_n \bigr).\)
Transition dynamics
\(\Pr(z_{n,t}=j\mid z_{n,t-1}=k,\;x^{A}_{n,t})= \operatorname{softmax}_j\!\bigl( \log A_{\text{base},kj}+W_{A,kj}\cdot x^{A}_{n,t} \bigr).\)
Emission
\(y_{n,t}\mid z_{n,t}=k\sim\text{Poisson}(\lambda_k).\)
Guide (variational family)
Point-mass approximation:
\(q(\pi_{\text{base}})=\delta(\pi_{\text{base}}-\hat\pi)\),
\(q(A_{\text{base}})=\delta(A_{\text{base}}-\hat A)\).
All \(\lambda\), \(W_\pi\), \(W_A\) are deterministic pyro.params and they are optimized during the training. Discrete states \(z_{n,t}\) are enumerated exactly, allowing all possible configurations to be calculated using TraceEnum_ELBO (without stochastic approximation).
Training setup
Loss: TraceEnum_ELBO(max_plate_nesting = 1)
Optimizer: Adam(lr = 2·10^{-2})
The learned parameters \(\hat\pi,\hat A,W_\pi,W_A,\hat\lambda\) show how the covariates influence entry probabilities, transition behaviour and expected donation rates.
K = 3
C_pi = 2 # birth_year_norm , gender_code
C_A = 2 # ages_norm , covid_years
# a priori asimmetriche
alpha_pi = torch.tensor([5., 2., 1.])
alpha_A = torch.tensor([[6.,1.,1.],
[1.,6.,1.],
[1.,1.,6.]])
@config_enumerate
def model(obs, x_pi, x_A):
N, T = obs.shape
# 1) Poisson rates
rates = pyro.param("rates",
0.5*torch.ones(K),
constraint=dist.constraints.positive)
# 2) Dirichlet priors
pi_base = pyro.sample("pi_base", dist.Dirichlet(alpha_pi))
A_base = pyro.sample("A_base",
dist.Dirichlet(alpha_A).to_event(1)) # shape (K,K)
log_pi_base = pi_base.log() # (K,)
log_A_base = A_base.log() # (K,K)
# 3) slope coefficients for covariates
W_pi = pyro.param("W_pi", torch.zeros(K, C_pi))
W_A = pyro.param("W_A", torch.zeros(K, K, C_A))
with pyro.plate("seqs", N):
# T=0
logits0 = log_pi_base + (x_pi @ W_pi.T) # (N,K)
z_prev = pyro.sample("z_0",
dist.Categorical(logits=logits0),
infer={"enumerate": "parallel"})
pyro.sample("y_0", dist.Poisson(rates[z_prev]), obs=obs[:,0])
# T>0
for t in range(1, T):
x_t = x_A[:, t, :] # (N,2)
logitsT = (log_A_base[z_prev] +
(W_A[z_prev] * x_t[:,None,:]).sum(-1))
z_t = pyro.sample(f"z_{t}",
dist.Categorical(logits=logitsT),
infer={"enumerate": "parallel"})
pyro.sample(f"y_{t}", dist.Poisson(rates[z_t]), obs=obs[:,t])
z_prev = z_t
# guide for the optimization
def guide(obs, x_pi, x_A):
# pi base
pi_q = pyro.param(
"pi_base_map",
torch.tensor([0.6, 0.3, 0.1]),
constraint=dist.constraints.simplex
)
# A base
A_init = torch.eye(K) * (K - 1.) + 1. # helps the diagonal
A_init = A_init / A_init.sum(-1, keepdim=True) # softmax, x>o and sum(x)=1
A_q = pyro.param(
"A_base_map",
A_init,
constraint=dist.constraints.simplex # x>o and sum(x)=1
)
# fix new pi_base and A_base for the next training iteration
# Delta is a trick to fix this values with sample
pyro.sample("pi_base", dist.Delta(pi_q).to_event(1))
pyro.sample("A_base", dist.Delta(A_q ).to_event(2))
# training
# pyro.clear_param_store()
# svi = SVI(model, guide,
# Adam({"lr": 2e-2}),
# loss=TraceEnum_ELBO(max_plate_nesting=1))
# for step in range(800):
# loss = svi.step(obs_torch, cov_init_torch, cov_tran_torch)
# if step % 200 == 0:
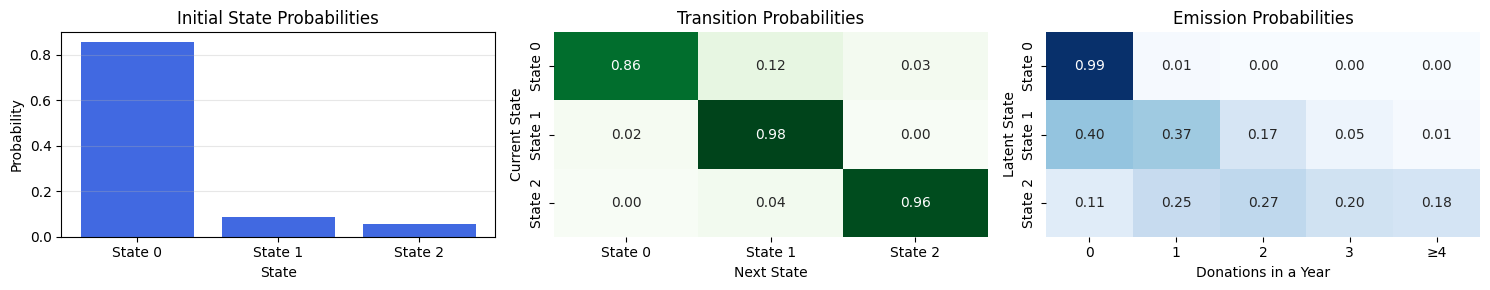
# print(f"{step:4d} ELBO = {loss:,.0f}")ELBO steadily decreases from \(\approx 181\) k to \(\approx 125\) k and then plateaus → optimisation has mostly converged.
Initial-state distribution \(\pi\) – State 0 dominates (85 %), followed by state 2 (8 %); state 1 is rare (7 %). Most donors start in state 0.
Transition matrix \(A\)
Poisson rates
Interpretation: the model has discovered three very stable behavioural profiles -non-donors, heavy donors, and light donors- with rare transitions between them.
def plot_hmm_params(transitions, initial_probs, emissions,
state_names=None, emission_names=None):
"""
plot in one row
- emission matrix
- initial probs
- transition matrix
"""
S = len(initial_probs)
K = emissions.shape[1]
if state_names is None:
state_names = [f"State {i}" for i in range(S)]
if emission_names is None:
emission_names = [str(i) for i in range(K)]
fig, axs = plt.subplots(1, 3, figsize=(15, 3))
# Initial probabilities
axs[0].bar(np.arange(S), initial_probs, color='royalblue')
axs[0].set_title('Initial State Probabilities')
axs[0].set_xlabel('State')
axs[0].set_ylabel('Probability')
axs[0].set_xticks(np.arange(S))
axs[0].set_xticklabels(state_names)
axs[0].grid(axis='y', alpha=0.3)
# Transition matrix
sns.heatmap(transitions, annot=True, fmt=".2f", cmap='Greens',
xticklabels=state_names, yticklabels=state_names, ax=axs[1], cbar=False)
axs[1].set_title('Transition Probabilities')
axs[1].set_xlabel('Next State')
axs[1].set_ylabel('Current State')
# Emission probabilities/matrix
sns.heatmap(emissions, annot=True, fmt=".2f", cmap='Blues',
xticklabels=emission_names, yticklabels=state_names, ax=axs[2], cbar=False)
axs[2].set_title('Emission Probabilities')
axs[2].set_xlabel('Donations in a Year')
axs[2].set_ylabel('Latent State')
plt.tight_layout()
plt.show()
def build_emission_matrix_truncated_poisson(rates, max_k=4):
S = len(rates)
K = max_k + 1
emissions = np.zeros((S, K))
for s in range(S):
for k in range(max_k):
emissions[s, k] = poisson.pmf(k, rates[s])
# it's a truncated poisson on the right side
emissions[s, max_k] = 1 - poisson.cdf(max_k-1, rates[s])
return emissions
import hmm_model
K = 3
W_pi, W_A, pi_base, A_base, lam = hmm_model.load_hmm_params(here("models\\hmm_full.pt"))
# covariate means (population averages)
x_mean_pi = cov_init_torch.mean(0).cpu().numpy()
x_mean_A = cov_tran_torch.mean((0,1)).cpu().numpy()
# mean initial probs and mean transition matrix
def softmax_row(v):
e = np.exp(v - v.max(-1, keepdims=True))
return e / e.sum(-1, keepdims=True)
# initial probabilities
logits_pi = np.log(pi_base) + W_pi @ x_mean_pi
pi_mean = softmax_row(logits_pi)
# transition matrix
A_mean = np.zeros((K, K))
for k in range(K):
logits = np.log(A_base[k]) + W_A[k] @ x_mean_A
A_mean[k] = softmax_row(logits)
def build_emission_matrix_trunc_poisson(rates, max_k=4):
S, G = len(rates), max_k + 1
M = np.zeros((S, G))
for s in range(S):
for k in range(max_k):
M[s, k] = poisson.pmf(k, rates[s])
M[s, max_k] = 1 - poisson.cdf(max_k - 1, rates[s])
return M
emissions_matrix = build_emission_matrix_trunc_poisson(lam, 4)
plot_hmm_params(
transitions = A_mean,
initial_probs = pi_mean,
emissions = emissions_matrix,
emission_names = [str(i) for i in range(4)] + ["≥4"]
)
Viterbi decoder
Goal: for each donor find the MAP latent path \(z_{0:T}^\ast\).
Plug-in parameters (posterior means) \[\hat\pi_k = \frac{\alpha_{\pi,k}}{\sum_{j}\alpha_{\pi,j}},\qquad \hat A_{kj} = \frac{\alpha_{A_{k j}}}{\sum_{j'}\alpha_{A_{k j'}}},\qquad \hat\lambda_k = \frac{\alpha_k}{\beta_k}.\]
Dynamic programming - Initial step \[\delta_0(k)=\log\hat\pi_k+\log\text{Poisson}(y_0\mid\hat\lambda_k).\]
Recursion for \(t=1,\dots,T\) \[\delta_t(j)=\max_k\bigl[\delta_{t-1}(k)+\log\hat A_{k j}\bigr]
+\log\text{Poisson}(y_t\mid\hat\lambda_j),\]
\[\psi_t(j)=\arg\max_k\bigl[\delta_{t-1}(k)+\log\hat A_{k j}\bigr].\]
Back-tracking Start with \(z_T^\ast=\arg\max_k\delta_T(k)\), then \(z_{t-1}^\ast=\psi_t(z_t^\ast)\) for \(t=T,\dots,1\).
def log_softmax_logits(logits, dim=-1):
return logits - logits.logsumexp(dim, keepdim=True)
def viterbi_paths_cov(obs, x_pi, x_A, K=3):
with torch.no_grad():
N, T = obs.shape
# ── learnt parameters ───────────────────────────────────────
lam = pyro.param("rates") # (K,)
W_pi = pyro.param("W_pi") # (K,2)
W_A = pyro.param("W_A") # (K,K,2)
pi_base = pyro.param("pi_base_map") # (K,) simplex
A_base = pyro.param("A_base_map") # (K,K) righe simplex
log_pi_base = pi_base.log() # (K,)
log_A_base = A_base.log() # (K,K)
# ── emission log-prob P(y_t | z_t=k) ───────────────────────
emis_log = torch.stack(
[dist.Poisson(l).log_prob(obs) for l in lam] # (K,N,T)
).permute(1, 2, 0) # (N,T,K)
# ── inizializzazione delta (t=0) ───────────────────────────
logits0 = log_pi_base + x_pi @ W_pi.T # (N,K)
log_pi = log_softmax_logits(logits0) # normalizza
delta = log_pi + emis_log[:, 0] # (N,K)
psi = torch.zeros(N, T, K, dtype=torch.long)
# ── forward pass t = 1 … T-1 ───────────────────────────────
for t in range(1, T):
x_t = x_A[:, t, :] # (N,2)
# logits[n, k_prev, k_next] = log A_base[k_prev,k_next] +
# W_A[k_prev,k_next] · x_t
base = log_A_base.unsqueeze(0) # (1,K,K)
slope = (W_A.unsqueeze(0) * x_t[:, None, None, :]).sum(-1)
logits = base + slope # (N,K,K)
log_A = log_softmax_logits(logits, dim=2)
score, idx = (delta.unsqueeze(2) + log_A).max(1) # max prev
psi[:, t] = idx
delta = score + emis_log[:, t]
# ── back-tracking ──────────────────────────────────────────
paths = torch.empty(N, T, dtype=torch.long)
last_state = delta.argmax(1)
paths[:, -1] = last_state
for t in range(T - 1, 0, -1):
last_state = psi[torch.arange(N), t, last_state]
paths[:, t-1] = last_state
return paths
paths = viterbi_paths_cov(obs_torch,
cov_init_torch, # (N,2)
cov_tran_torch, # (N,T,2)
K=3)
switch_rate = (paths[:, 1:] != paths[:, :-1]).any(1).float().mean()
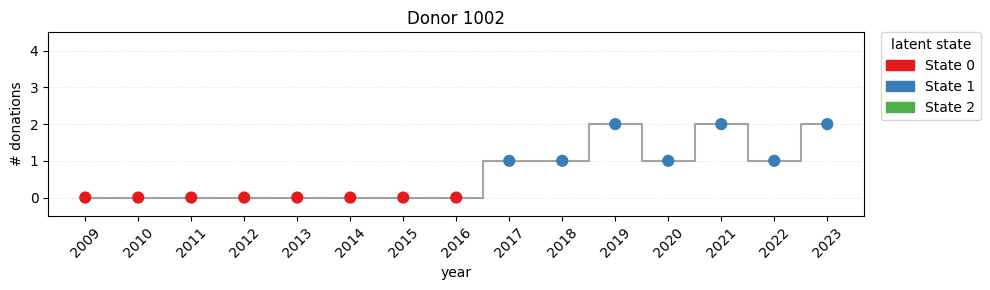
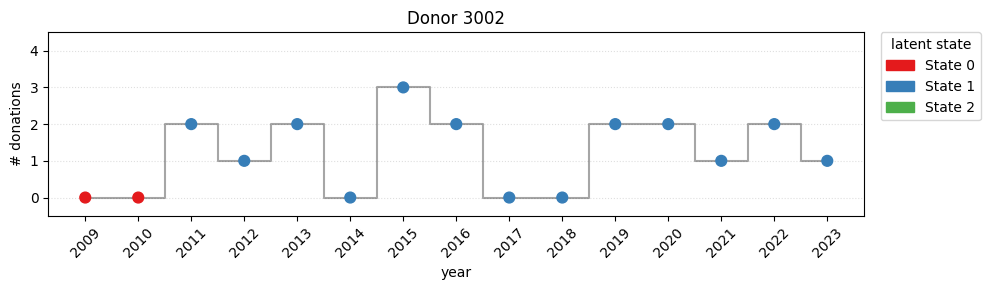
print(f"switch rate = {switch_rate:.2%}")switch rate = 78.52%Looking the plot, it’s easy to detect a strong movement from state 0 to state 1 during the covid years.
counts = np.apply_along_axis(lambda col: np.bincount(col, minlength=3),
0, paths) # (K,T)
props = counts / counts.sum(0, keepdims=True)
plt.figure(figsize=(8,4))
for k,c in enumerate(['tab:orange','tab:blue','tab:green']):
plt.plot(props[k], label=f'state {k}', color=c)
plt.xlabel('year index'); plt.ylabel('population share')
plt.title('State occupancy over time'); plt.legend(); plt.tight_layout()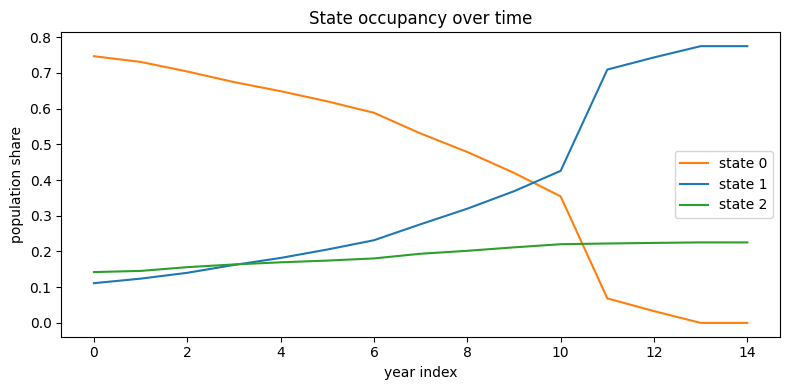
Pick a few donors and overlay observations + decoded state for an easy interpretation of the model.
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap, BoundaryNorm
from matplotlib.patches import Patch
K = 3
state_cols = ['#e41a1c', '#377eb8', '#4daf4a']
cmap = ListedColormap(state_cols)
norm = BoundaryNorm(np.arange(-0.5, K+0.5, 1), cmap.N)
years_axis = np.arange(2009, 2024)
yticks_vals = np.arange(0, 5)
def plot_one(idx):
x = obs_torch[idx].cpu().numpy()
z = paths[idx]
T = len(x)
assert T == len(years_axis), "years_axis length must match T"
plt.figure(figsize=(10, 3))
plt.scatter(range(T), x, c=z, cmap=cmap, norm=norm, s=60, zorder=3)
plt.step(range(T), x, where='mid', color='k', alpha=.35, zorder=2)
plt.xticks(ticks=range(T), labels=years_axis, rotation=45)
plt.yticks(ticks=yticks_vals)
plt.ylim(-0.5, 4.5) # blocca a 0–4
plt.grid(axis='y', linestyle=':', alpha=.4, zorder=1)
handles = [Patch(color=state_cols[k], label=f'State {k}') for k in range(K)]
plt.legend(handles=handles, title='latent state',
bbox_to_anchor=(1.02, 1), loc='upper left', borderaxespad=0.)
plt.title(f'Donor {idx}')
plt.xlabel('year')
plt.ylabel('# donations')
plt.tight_layout()
plt.show()
for i in [100, 1002, 3002]:
plot_one(i)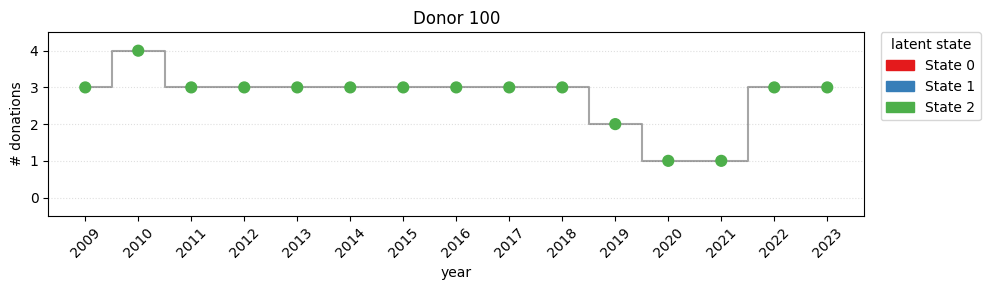
def plot_W_pi_heat(W_pi):
sns.heatmap(W_pi,
annot=True, fmt=".2f",
xticklabels=cov_names_pi,
yticklabels=[f"S{k}" for k in range(W_pi.shape[0])],
cmap="coolwarm", center=0)
plt.title("W_pi – slopes on log π")
plt.tight_layout(); plt.show()
def plot_W_A_heat(W_A):
K, _, C_A = W_A.shape
fig, axes = plt.subplots(K, K,
figsize=(C_A*1.6, K*1.6),
sharex=True, sharey=True)
vmin, vmax = W_A.min(), W_A.max()
for i in range(K):
for j in range(K):
ax = axes[i, j]
mat = W_A[i, j].reshape(1, -1)
sns.heatmap(mat, ax=ax,
vmin=vmin, vmax=vmax,
cmap="coolwarm", cbar=False,
xticklabels=cov_names_A,
yticklabels=[])
ax.set_title(f"{i}→{j}", fontsize=8)
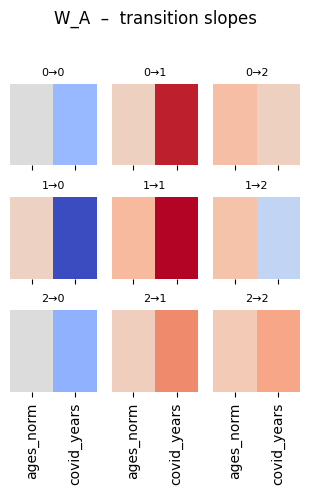
plt.suptitle("W_A – transition slopes", y=1.02)
plt.tight_layout(); plt.show()
plot_W_pi_heat(W_pi)
plot_W_A_heat(W_A)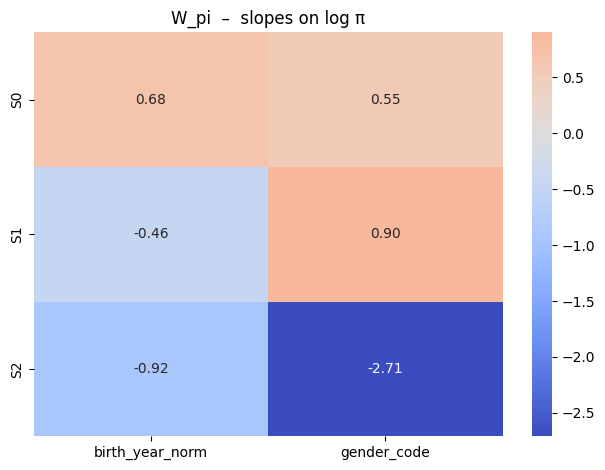
# Compute the (mean , std) that were used for z-scoring
# birth-year
birth_year_orig = df["birth_year"].to_numpy()
birth_year_mean = birth_year_orig.mean()
birth_year_std = birth_year_orig.std()
# ages (matrix N × T built from birth_year and calendar years)
years_num = np.array([int(c[2:]) for c in sorted([c for c in df.columns
if c.startswith("y_")])])
ages = years_num[None, :] - df["birth_year"].to_numpy()[:, None]
ages_mean = ages.mean()
ages_std = ages.std()
# Store them in the 'stats' dictionary
stats = {
"birth_year_norm": (birth_year_mean, birth_year_std),
"ages_norm": (ages_mean, ages_std)
}
def softmax(v: np.ndarray) -> np.ndarray:
v = v - np.max(v)
e = np.exp(v)
return e / np.sum(e)
def original_values(var_name: str) -> np.ndarray:
"""
Return a 1-D NumPy array containing the covariate on
its natural scale (no z-scoring).
"""
if var_name == "birth_year_norm":
return df["birth_year"].to_numpy()
if var_name == "ages_norm":
return ages.reshape(-1)
if var_name == "gender_code":
return np.array([0., 1.]) # binary
if var_name == "covid_years":
return np.array([0., 1.]) # binary
raise ValueError(f"Unknown covariate {var_name}")
# mean / std used when the variable was z-scored ----------------
stats = {
"birth_year_norm": (birth_year_mean, birth_year_std),
"ages_norm": (ages_mean, ages_std)
}
def to_norm(var_name: str, x_orig: np.ndarray) -> np.ndarray:
"""
Convert an array of ORIGINAL values to the z-scored scale
expected by the model.
"""
if var_name in stats:
mu, sd = stats[var_name]
return (x_orig - mu) / sd
return x_orig
def plot_pi_vs_cov_orig(var="birth_year_norm", grid_orig=None):
if var not in cov_names_pi:
raise ValueError(f"{var} is not a π covariate")
idx = cov_names_pi.index(var)
col_orig = original_values(var)
if grid_orig is None:
uniq = np.unique(col_orig)
grid_orig = uniq if len(uniq) <= 3 else np.linspace(col_orig.min(),
col_orig.max(), 41)
x_ref_norm = cov_init.mean(0) # (2,)
curves = []
for v_orig in grid_orig:
x_norm = x_ref_norm.copy()
x_norm[idx] = to_norm(var, v_orig)
logits = logits_pi + W_pi @ x_norm # (K,)
curves.append(softmax(logits))
curves = np.vstack(curves) # (G, K)
for k, c in enumerate(state_cols):
plt.plot(grid_orig, curves[:, k], color=c, label=f"state {k}")
plt.xlabel(var.replace("_norm", "")); plt.ylabel("πₖ(x)")
plt.title(f"Initial-state probability vs {var.replace('_norm', '')}")
plt.legend(); plt.grid(ls=":"); plt.show()
plot_pi_vs_cov_orig("birth_year_norm")
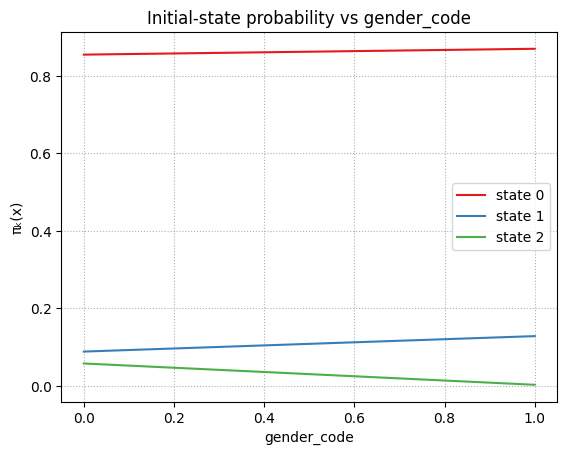
plot_pi_vs_cov_orig("gender_code")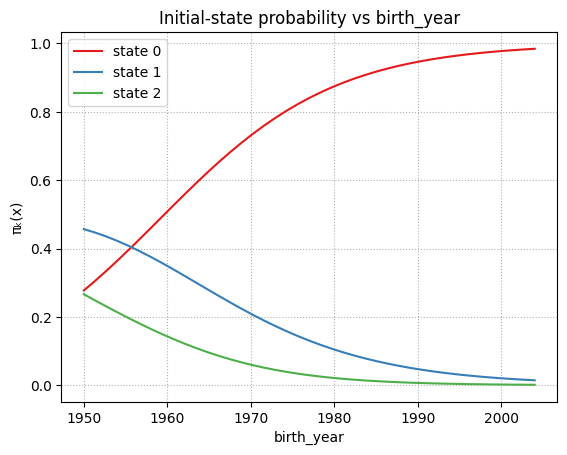
def _softmax_np(v):
v = v - np.max(v, axis=-1, keepdims=True)
e = np.exp(v)
return e / np.clip(e.sum(axis=-1, keepdims=True), 1e-30, None)
def plot_trans_simple(var,
prev_state,
cov_names_A,
xA_ref, # (C_A,) vettore di riferimento
W_A, # (K, K, C_A)
A_base, # (K, K) (row-simplex)
grid=None,
colors=None):
"""
Traccia P(z_t=j | z_{t-1}=prev_state, x_A[var]=v) al variare di v,
mantenendo tutte le altre covariate fissate a xA_ref.
Tutto è in scala modello
"""
K, _, C_A = W_A.shape
idx = cov_names_A.index(var)
xA_ref = np.asarray(xA_ref, dtype=float).ravel().copy()
assert xA_ref.shape[0] == C_A, "xA_ref deve essere (C_A,)"
# if grid is None:
# v0 = xA_ref[idx]
# grid = np.linspace(v0 - 2.0, v0 + 2.0, 41)
# if colors is None:
# base = ['#e41a1c','#377eb8','#4daf4a','#984ea3','#ff7f00',
# '#ffff33','#a65628','#f781bf','#999999']
# colors = (base * ((K + len(base) - 1)//len(base)))[:K]
logA0 = np.log(np.clip(A_base, 1e-30, None)) # (K,K)
probs = np.zeros((len(grid), K))
for g, val in enumerate(grid):
x_vec = xA_ref.copy()
x_vec[idx] = float(val)
logits = logA0[prev_state] + (W_A[prev_state] @ x_vec) # (K,)
probs[g] = _softmax_np(logits[None, :]).ravel()
for j in range(K):
plt.plot(grid, probs[:, j], color=colors[j], label=f"{prev_state}→{j}")
plt.xlabel(var); plt.ylabel("transition prob.")
plt.title(f"Transition from state {prev_state} vs {var}")
plt.grid(ls=":"); plt.legend(); plt.tight_layout(); plt.show()
def expected_y_simple(var,
cov_names_pi,
xpi_ref, # (C_pi,) vettore di riferimento (es. media su N)
W_pi, # (K, C_pi)
pi_base, # (K,) simplex
lam_k, # (K,) tasso Poisson per stato (pre-calcolato)
grid=None):
"""
Traccia E[y0 | x_pi[var]=v] al variare di v,
mantenendo le altre covariate fissate a xpi_ref.
"""
K, C_pi = W_pi.shape
idx = cov_names_pi.index(var)
xpi_ref = np.asarray(xpi_ref, dtype=float).ravel().copy()
assert xpi_ref.shape[0] == C_pi, "xpi_ref deve essere (C_pi,)"
# if grid is None:
# v0 = xpi_ref[idx]
# grid = np.linspace(v0 - 2.0, v0 + 2.0, 41)
log_pi0 = np.log(np.clip(np.asarray(pi_base, dtype=float).ravel(), 1e-30, None))
lam_k = np.asarray(lam_k, dtype=float).ravel()
assert lam_k.shape[0] == K, "lam_k deve essere (K,)"
y_exp = np.zeros(len(grid))
for g, val in enumerate(grid):
x_vec = xpi_ref.copy()
x_vec[idx] = float(val)
logits = log_pi0 + (W_pi @ x_vec) # (K,)
pi_x = _softmax_np(logits[None, :]).ravel()
y_exp[g] = float((pi_x * lam_k).sum())
plt.plot(grid, y_exp, "-o", ms=3)
plt.xlabel(var); plt.ylabel("E[y0 | x]")
plt.title(f"Expected count at t=0 vs {var}")
plt.grid(ls=":"); plt.tight_layout(); plt.show()
xA_ref = cov_tran_torch.mean(dim=(0,1)).detach().cpu().numpy() # (C_A,)
plot_trans_simple("covid_years", prev_state=1, cov_names_A=cov_names_A,
xA_ref=xA_ref, W_A=W_A, A_base=A_base, grid=np.array([0.,1.]))
xpi_ref = cov_init_torch.mean(0).detach().cpu().numpy() # (C_pi,)
expected_y_simple("gender_code", cov_names_pi, xpi_ref, W_pi, pi_base, lam, grid=np.array([0.,1.]))--------------------------------------------------------------------------- TypeError Traceback (most recent call last) Cell In[13], line 90 87 plt.grid(ls=":"); plt.tight_layout(); plt.show() 89 xA_ref = cov_tran_torch.mean(dim=(0,1)).detach().cpu().numpy() # (C_A,) ---> 90 plot_trans_simple("covid_years", prev_state=1, cov_names_A=cov_names_A, 91 xA_ref=xA_ref, W_A=W_A, A_base=A_base, grid=np.array([0.,1.])) 93 xpi_ref = cov_init_torch.mean(0).detach().cpu().numpy() # (C_pi,) 94 expected_y_simple("gender_code", cov_names_pi, xpi_ref, W_pi, pi_base, lam, grid=np.array([0.,1.])) Cell In[13], line 44, in plot_trans_simple(var, prev_state, cov_names_A, xA_ref, W_A, A_base, grid, colors) 41 probs[g] = _softmax_np(logits[None, :]).ravel() 43 for j in range(K): ---> 44 plt.plot(grid, probs[:, j], color=colors[j], label=f"{prev_state}→{j}") 45 plt.xlabel(var); plt.ylabel("transition prob.") 46 plt.title(f"Transition from state {prev_state} vs {var}") TypeError: 'NoneType' object is not subscriptable
# Heat-map helper: original-scale grids for the A–slopes model
def original_values(var: str) -> np.ndarray:
"""
Return a 1-D NumPy array with the covariate on its
natural (unnormalised) scale.
"""
if var == "birth_year_norm":
return df["birth_year"].to_numpy()
if var == "ages_norm":
return ages.reshape(-1)
if var == "gender_code":
return np.array([0., 1.]) # binary
if var == "covid_years":
return np.array([0., 1.]) # binary
raise ValueError(f"Unknown covariate {var}")
def to_norm(var: str, x_orig: np.ndarray) -> np.ndarray:
"""Convert ORIGINAL values to the z-scored scale."""
if var in stats:
mu, sd = stats[var]
return (x_orig - mu) / sd
return x_orig
def plot_transition_grid_orig(var_name: str,
cov_names: list,
log_A0: np.ndarray, # (K , K )
W_A: np.ndarray, # (K , K , C)
full_cov: np.ndarray, # (N , T , C) normalised
grid_vals_orig=None,
n_grid: int = 21):
K, _, C = W_A.shape
idx = cov_names.index(var_name)
if grid_vals_orig is None:
uniq = np.unique(original_values(var_name))
if len(uniq) <= 2: # binary / categorical
grid_vals_orig = uniq.astype(float)
plot_mode = "bar"
else: # continuous
lo, hi = uniq.min(), uniq.max()
grid_vals_orig = np.linspace(lo, hi, n_grid)
plot_mode = "line"
else:
plot_mode = "line"
grid_vals_norm = to_norm(var_name, grid_vals_orig)
x_mean = full_cov.mean(axis=(0, 1)) # (C,)
mats = np.empty((len(grid_vals_norm), K, K)) # (G , K , K)
for g, v_norm in enumerate(grid_vals_norm):
x_vec = x_mean.copy()
x_vec[idx] = v_norm
logits = log_A0 + np.tensordot(W_A, x_vec, axes=([2], [0]))
probs = np.exp(logits)
probs /= probs.sum(axis=1, keepdims=True) # row soft-max
mats[g] = probs
fig, axes = plt.subplots(K, K,
figsize=(K * 2.4, K * 2.4),
sharex=True, sharey=True)
for i in range(K):
for j in range(K):
ax = axes[i, j]
if plot_mode == "line":
ax.plot(grid_vals_orig, mats[:, i, j], color="steelblue")
else: # bar plot (binary)
ax.bar(grid_vals_orig, mats[:, i, j],
width=0.6, color=['#1f77b4', '#ff7f0e'])
ax.set_xticks(grid_vals_orig)
ax.set_title(f"P({i}→{j})", fontsize=8)
if i == K - 1:
ax.set_xlabel(var_name.replace("_norm", ""))
if j == 0:
ax.set_ylabel("Prob")
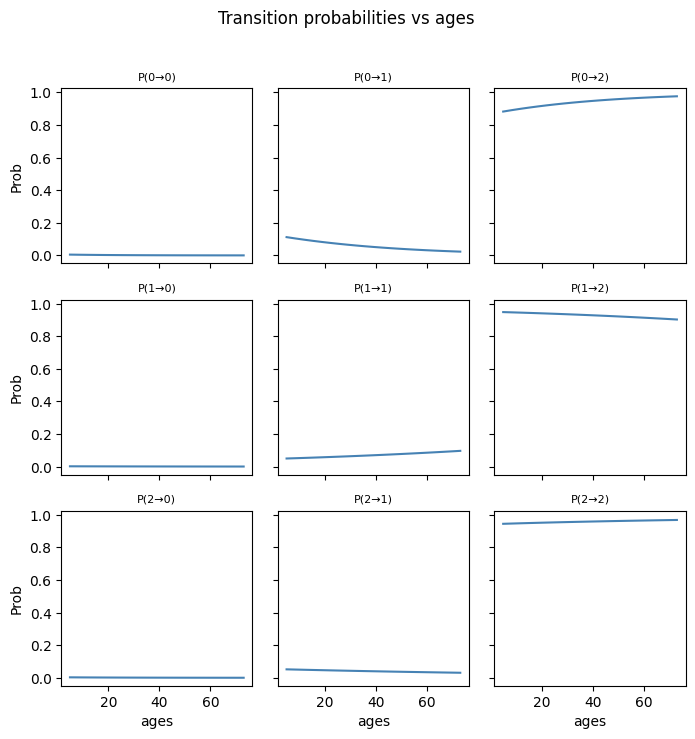
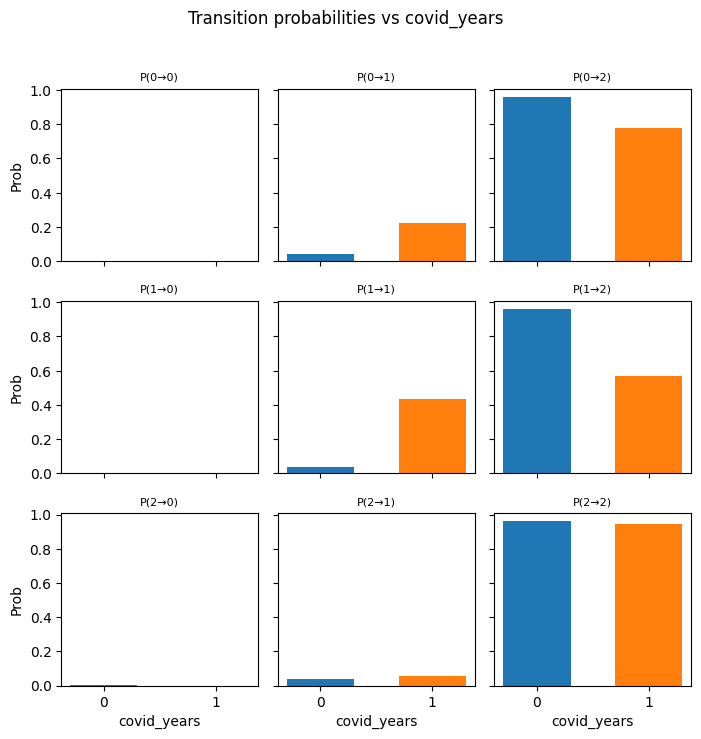
plt.suptitle(f"Transition probabilities vs "
f"{var_name.replace('_norm', '')}",
y=1.02)
plt.tight_layout()
plt.show()
cov_names_A = ["ages_norm", "covid_years"]
full_cov_A = cov_tran
for var in cov_names_A:
plot_transition_grid_orig(var,
cov_names_A,
logits,
W_A,
full_cov_A)